Atomic Force Microscopy ( AFM ) can be utilized to determine the mechanical properties of tumor tissues in different kinds of cancers, for example breast cancer, liver cancer and lung cancer.
Oral squamous cell carcinoma (OSCC) is a common subtype of head and neck and other malignant tumors that occurs in increasing numbers. It is therefore important to learn more about the biological factors connected with the early diagnosis and treatment of OSCC. *
The human trophoblast cell surface antigen 2 (TROP2), which is also called tumor-associated calcium signal transduction-2 (TACSTD-2), is a surface glycoprotein encoded by TACSTD. *
Among the various biochemical mechanisms involved in tumorigenesis, the role of β-catenin has been studied extensively. This has shed light on the biological functions of TROP2 and its use as a prognostic biomarker for OSCC. *
TROP2 regulates tumorigenic properties including cancer cell adhesion, invasion, and migration and is overexpressed in many human cancers. Inhibiting TROP2 expression has shown promise in clinical applications. *
In the article “Tissue mechanics and expression of TROP2 in oral squamous cell carcinoma with varying differentiation” Baoping Zhang, Shuting Gao, Ruiping Li, Yiting Li, Rui Cao, Jingyang Cheng, Yumeng Guo, Errui Wang, Ying Huang and Kailiang Zhang investigate the role of TROP2 in OSCC patients using a combination of biophysical approaches including atomic force microscopy. *
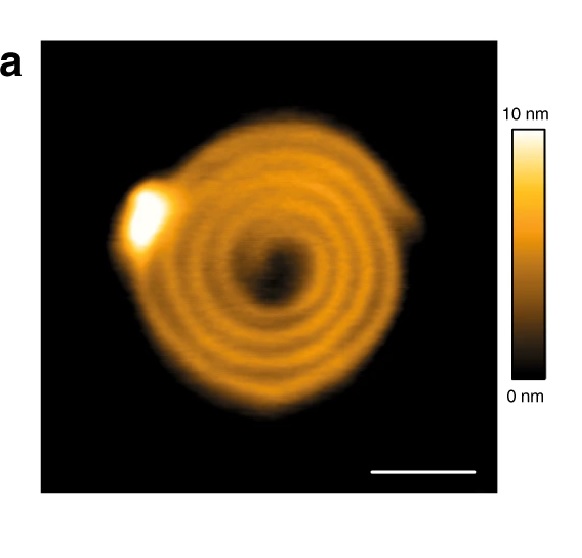
The authors demonstrate the tissue morphology and mechanics of OSCC samples during tumor development using NanoWorld Pointprobe® CONTR AFM probes for the Atomic Force Microscopy described in the article and they believe that their findings will help develop TROP2 in accurately diagnosing OSCC in tumors with different grades of differentiation. *

Surface morphology of OSCC tissue sections via AFM detection, irregular morphology appeared in the low differentiation
*Baoping Zhang, Shuting Gao, Ruiping Li, Yiting Li, Rui Cao, Jingyang Cheng, Yumeng Guo, Errui Wang, Ying Huang and Kailiang Zhang
Tissue mechanics and expression of TROP2 in oral squamous cell carcinoma with varying differentiation
BMC Cancer volume 20, Article number: 815 (2020)
DOI: https://doi.org/10.1186/s12885-020-07257-7
Please follow this external link to read the whole article: https://rdcu.be/cfC9G
Open Access : The article “Tissue mechanics and expression of TROP2 in oral squamous cell carcinoma with varying differentiation” by Baoping Zhang, Shuting Gao, Ruiping Li, Yiting Li, Rui Cao, Jingyang Cheng, Yumeng Guo, Errui Wang, Ying Huang and Kailiang Zhang is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/.